How can computers help crops grow better?
Computers have revolutionised the planet in many ways. Plant science is one of the fields to have benefitted from recent advances in technology, thanks to computer scientists such as Professor Tony Pridmore at the University of Nottingham, UK, who are using their computing skills to address challenges in plant biology. Tony is the director of PhenomUK, the UK’s plant phenotyping network, where new software and imaging techniques developed by computer scientists and engineers, in collaboration with plant scientists, could improve crop yields around the world
TALK LIKE A PLANT PHENOTYPING RESEARCHER
CANOPY CLOSURE – a measure of plant density in an area
COMPUTER VISION – a branch of computer science aimed at extracting information about a viewed scene from images
GENOTYPE – the genetic makeup of an organism
MACHINE LEARNING – a computer program that can teach itself to perform a task
MAGNETIC RESONANCE IMAGING (MRI) – an imaging technique using magnetic fields to view the internal structure of an organism
PHENOTYPE – the observable physical properties of an organism resulting from its genotype and the environment it inhabits
PLANT PHENOTYPING – the assessment of plant traits
PLANT TRAITS – any feature of a plant that is measurable (e.g., growth rate, drought tolerance) from a cellular to organism level
With an ever-increasing population, the world needs more food. Yet climate change and ecological destruction mean food must be produced in increasingly hostile environments. We therefore need crops that are not only more productive but are also productive in harsher conditions, with significantly less or significantly more water than before, for example.
Plant phenotyping is changing the way crops are developed by assessing how plants grow in different environments. Images of plants are captured and converted into data that can be statistically analysed, allowing researchers to determine which plants are most suitable for which conditions. While plant scientists are needed to conduct plant experiments and interpret the results, the process of converting images into data requires computer scientists like Professor Tony Pridmore. A professor of computer science and head of the Computer Vision Laboratory at the University of Nottingham, Tony is also director of PhenomUK, the plant phenotyping network that aims to address issues in plant biology by bringing computer scientists and engineers into the field.
PLANT PHENOTYPING METHODS
The phenotype of a plant is defined by its characteristics and traits, which are controlled by interactions between the plant’s genotype and its growing environment. “Plant phenotyping is about measuring plants in an objective and quantitative way, by measuring the structural and functional properties of a large number of plants,” explains Tony. “How big are they? What shape are they? How fast are they growing? How do they react to drought, pests and various stresses?”
Phenotyping researchers grow plants in different experimental environments, from climate-controlled growth chambers in labs, to greenhouses and, finally, to open fields. To study them, these plants are imaged with digital cameras viewing the plants at different scales depending on what the researchers are interested in, from a whole field to an individual plant to a plant organ such as a single leaf. The large number of images required means data collection and analysis are usually automated.
“Different technologies are used to take images in different environments,” explains Tony. “In growth chambers, conditions are artificial so a camera can be placed anywhere.” Imaging becomes more complex in greenhouses, as other objects interfere with the plant of interest, while in fields, wind and rain cause plants to move which adds further challenges. “Cameras taking these images may be in a fixed position, or mounted on moving platforms, drones or automated vehicles,” Tony says.
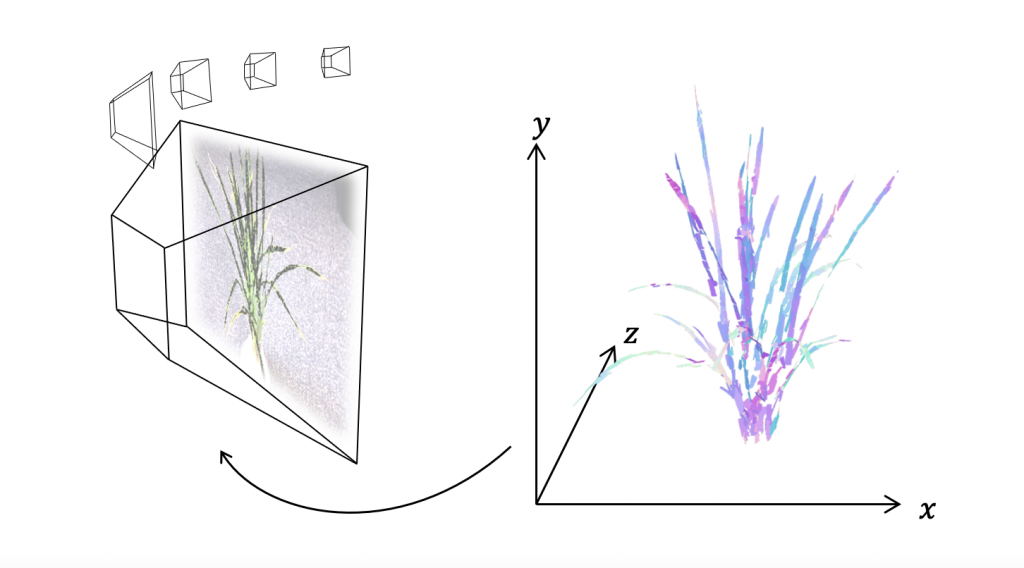
As well as standard camera images, researchers also take hyperspectral images which detect features outside the range of visible light. “Hyperspectral photos can be split into hundreds of images, all showing different things, such as features visible in ultraviolet,” explains Tony. The interior of plants can also be imaged, with X-ray images providing information about the plant’s internal structure and magnetic resonance imaging (MRI) showing how fluids move through the plant. Standard colour images can be used to build 3D models of the plant if multiple photos are taken at different angles.
CONVERTING PLANT IMAGES INTO STATISTICAL DATA
To convert images of plants into useful data that can be statistically analysed requires skills from computer science, which is where Tony’s work in computer vision comes in. “An image is composed of pixels,” explains Tony, “and to extract data from an image usually involves segmenting the image by writing computer code to define what the different pixels represent. For example, we might define which pixels represent a leaf so the computer can separate out all the leaves from the background of an image.”
This code is no longer written by hand as machine learning means computers can now be taught to segment images and recognise objects. By marking all the leaves in a set of images and feeding them into a computer, the machine learning algorithm will develop its own rules to identify leaves in other images.
Once the computer has identified the features of the plants in the images, more code is written so the computer measures the characteristics of these features, such as leaf length, plant height or growth rate. With the images converted into numerical data, phenotyping researchers can then statistically analyse the properties of the plants. “We might do correlation analysis between genetic profiles and plants traits to determine which genes result in which physical traits in the plant,” says Tony.
PHENOMUK
A significant challenge for addressing global food production is that plant scientists do not often have sufficient computing skills to develop the tools they need to extract the required data from their plant experiments, while computer scientists do not have the biological knowledge necessary for answering plant-related research questions. PhenomUK aims to solve this problem by establishing collaborations between plant and computer scientists. Then, with their combined knowledge and expertise, they can find solutions to crop development together. “Plant phenotyping involves applying skills from computing to address issues in plant science,” Tony explains. “Our main goal is to build a phenotyping community by bringing computer scientists and engineers into contact with plant biologists.”
PhenomUK also funds research projects which have components from both plant and computer sciences. One project is probing the internal structure of plants using microwave radiation, a cheaper alternative to X-ray and MRI imaging. This could be used to detect abnormalities in the internal structure of fruit or visualise underground root structures. Another project uses drones to take high quality, high frequency aerial images of wheat crops to automatically measure plant height, canopy closure and leaf area.
A recently funded project focuses on speeding up plant development by using deep machine learning to predict the future growth of plants. If software can be built to predict what a plant’s roots will look like in the future, based on a series of images of the roots’ growth so far, then plants will not need to be grown for as long before useful data can be collected. Another project is developing a laser scanning device which can produce high resolution 3D models of plant features. This technique could replace current imaging techniques which struggle in outdoor growing environments due to changing light and wind conditions.
THE FUTURE OF PHENOTYPING
The equipment required for plant phenotyping, such as X-ray and MRI resources, is expensive. It would be uneconomical for every country to have their own equipment, which is why PhenomUK is closely related to EMPHASIS, a European research infrastructure that will share phenotyping facilities and data across Europe. This will allow researchers across the continent to contribute to the expanding field of plant phenotyping, enabling plant biologists and computer scientists to ensure crops will be able to feed the population of the future.
Reference
https://doi.org/10.33424/FUTURUM217
TALK LIKE A PLANT PHENOTYPING RESEARCHER
CANOPY CLOSURE – a measure of plant density in an area
COMPUTER VISION – a branch of computer science aimed at extracting information about a viewed scene from images
GENOTYPE – the genetic makeup of an organism
MACHINE LEARNING – a computer program that can teach itself to perform a task
MAGNETIC RESONANCE IMAGING (MRI) – an imaging technique using magnetic fields to view the internal structure of an organism
PHENOTYPE – the observable physical properties of an organism resulting from its genotype and the environment it inhabits
PLANT PHENOTYPING – the assessment of plant traits
PLANT TRAITS – any feature of a plant that is measurable (e.g., growth rate, drought tolerance) from a cellular to organism level
Plant phenotyping is changing the way crops are developed by assessing how plants grow in different environments. Images of plants are captured and converted into data that can be statistically analysed, allowing researchers to determine which plants are most suitable for which conditions. While plant scientists are needed to conduct plant experiments and interpret the results, the process of converting images into data requires computer scientists like Professor Tony Pridmore. A professor of computer science and head of the Computer Vision Laboratory at the University of Nottingham, Tony is also director of PhenomUK, the plant phenotyping network that aims to address issues in plant biology by bringing computer scientists and engineers into the field.
PLANT PHENOTYPING METHODS
The phenotype of a plant is defined by its characteristics and traits, which are controlled by interactions between the plant’s genotype and its growing environment. “Plant phenotyping is about measuring plants in an objective and quantitative way, by measuring the structural and functional properties of a large number of plants,” explains Tony. “How big are they? What shape are they? How fast are they growing? How do they react to drought, pests and various stresses?”
Phenotyping researchers grow plants in different experimental environments, from climate-controlled growth chambers in labs, to greenhouses and, finally, to open fields. To study them, these plants are imaged with digital cameras viewing the plants at different scales depending on what the researchers are interested in, from a whole field to an individual plant to a plant organ such as a single leaf. The large number of images required means data collection and analysis are usually automated.
“Different technologies are used to take images in different environments,” explains Tony. “In growth chambers, conditions are artificial so a camera can be placed anywhere.” Imaging becomes more complex in greenhouses, as other objects interfere with the plant of interest, while in fields, wind and rain cause plants to move which adds further challenges. “Cameras taking these images may be in a fixed position, or mounted on moving platforms, drones or automated vehicles,” Tony says.
As well as standard camera images, researchers also take hyperspectral images which detect features outside the range of visible light. “Hyperspectral photos can be split into hundreds of images, all showing different things, such as features visible in ultraviolet,” explains Tony. The interior of plants can also be imaged, with X-ray images providing information about the plant’s internal structure and magnetic resonance imaging (MRI) showing how fluids move through the plant. Standard colour images can be used to build 3D models of the plant if multiple photos are taken at different angles.
CONVERTING PLANT IMAGES INTO STATISTICAL DATA
To convert images of plants into useful data that can be statistically analysed requires skills from computer science, which is where Tony’s work in computer vision comes in. “An image is composed of pixels,” explains Tony, “and to extract data from an image usually involves segmenting the image by writing computer code to define what the different pixels represent. For example, we might define which pixels represent a leaf so the computer can separate out all the leaves from the background of an image.”
This code is no longer written by hand as machine learning means computers can now be taught to segment images and recognise objects. By marking all the leaves in a set of images and feeding them into a computer, the machine learning algorithm will develop its own rules to identify leaves in other images.
Once the computer has identified the features of the plants in the images, more code is written so the computer measures the characteristics of these features, such as leaf length, plant height or growth rate. With the images converted into numerical data, phenotyping researchers can then statistically analyse the properties of the plants. “We might do correlation analysis between genetic profiles and plants traits to determine which genes result in which physical traits in the plant,” says Tony.
PHENOMUK
A significant challenge for addressing global food production is that plant scientists do not often have sufficient computing skills to develop the tools they need to extract the required data from their plant experiments, while computer scientists do not have the biological knowledge necessary for answering plant-related research questions. PhenomUK aims to solve this problem by establishing collaborations between plant and computer scientists. Then, with their combined knowledge and expertise, they can find solutions to crop development together. “Plant phenotyping involves applying skills from computing to address issues in plant science,” Tony explains. “Our main goal is to build a phenotyping community by bringing computer scientists and engineers into contact with plant biologists.”
PhenomUK also funds research projects which have components from both plant and computer sciences. One project is probing the internal structure of plants using microwave radiation, a cheaper alternative to X-ray and MRI imaging. This could be used to detect abnormalities in the internal structure of fruit or visualise underground root structures. Another project uses drones to take high quality, high frequency aerial images of wheat crops to automatically measure plant height, canopy closure and leaf area.
A recently funded project focuses on speeding up plant development by using deep machine learning to predict the future growth of plants. If software can be built to predict what a plant’s roots will look like in the future, based on a series of images of the roots’ growth so far, then plants will not need to be grown for as long before useful data can be collected. Another project is developing a laser scanning device which can produce high resolution 3D models of plant features. This technique could replace current imaging techniques which struggle in outdoor growing environments due to changing light and wind conditions.
THE FUTURE OF PHENOTYPING
The equipment required for plant phenotyping, such as X-ray and MRI resources, is expensive. It would be uneconomical for every country to have their own equipment, which is why PhenomUK is closely related to EMPHASIS, a European research infrastructure that will share phenotyping facilities and data across Europe. This will allow researchers across the continent to contribute to the expanding field of plant phenotyping, enabling plant biologists and computer scientists to ensure crops will be able to feed the population of the future.
 PROFESSOR TONY PRIDMORE
PROFESSOR TONY PRIDMORE
Professor of Computer Science, Head of the Computer Vision Laboratory, University of Nottingham, UK
Director of PhenomUK
FIELD OF RESEARCH: Plant Phenotyping
RESEARCH PROJECT: Using computing methods to extract information about plants from images
FUNDER: UK Research and Innovation (UKRI), through the Technology Touching Life programme
 PROFESSOR TONY PRIDMORE
PROFESSOR TONY PRIDMORE
Professor of Computer Science, Head of the Computer Vision Laboratory, University of Nottingham, UK
Director of PhenomUK
FIELD OF RESEARCH: Plant Phenotyping
RESEARCH PROJECT: Using computing methods to extract information about plants from images
FUNDER: UK Research and Innovation (UKRI), through the Technology Touching Life programme
ABOUT PLANT PHENOTYPING
The plant science community urgently needs computer science expertise to develop the tools needed to collect and process the data required by plant biologists to ensure food security. Plant phenotyping is the crucial link for plant breeders to grow crops that produce high yields, can tolerate droughts and floods, and are resistant to pests and diseases.
Working in plant phenotyping allows computer scientists to apply their skills to address real-world problems. “A satisfying aspect of phenotyping work is that technical problems are interesting and need real developments in computer science to be made,” says Tony. The collaborations between research fields also offers huge scope for research innovation and discovery, though this interdisciplinary work can be challenging. “Explaining to plant scientists what we, the computer scientists, have done and how they should use the techniques can be challenging,” Tony says.
EXPLORE A CAREER IN PLANT PHENOTYPING
• For more information about plant phenotyping, visit the websites for PhenomUK (www.phenomuk.net) and the EMPHASIS project (emphasis.plant-phenotyping.eu).
• Most phenotyping researchers will work in academic institutions such as universities or research facilities. As a plant biologist, you could design experiments to assess the effect of different conditions on plant growth. As an engineer, you could build new equipment and imaging technology to collect images of the plants in these experiments. As a computer scientist, you could write code and develop software to convert these images into quantitative data. As a statistician, you could analyse the data extracted from these images to interpret the results of the experiments.
• With knowledge of plant phenotyping, you could also work in plant breeding, using the information provided by phenotyping researchers to develop more productive crops. The British Society of Plant Breeding has a webpage explaining careers in plant breeding: www.bspb.co.uk/plant-breeding/careers-in-plant-breeding
• You can enter the field of plant phenotyping from the computing/engineering or biology/plant science side. Knowledge of both sides is useful, but not essential. It is more important to have an interest in both.
• For computer science degrees, A-levels in maths, computing and physics are useful, though not always necessary. For biology, sciences like biology, chemistry or physics, as well as maths, are useful.
• During a computer science or engineering degree, a focus on artificial intelligence, data analysis or robotics would be useful. Likewise, for a biology degree, modules in computing would be important. The University of Nottingham offers a plant science degree that includes a year of computer science.
HOW DID TONY BECOME A PLANT PHENOTYPING RESEARCHER?
Computers weren’t easily available when I was growing up, so I wasn’t exposed to programming until I was well into my teens. Music has always been a huge interest, and my first career choice was to be a guitarist and bass player.
I grew up in a small town in Northamptonshire where all school pupils had to do a vocational course to prepare them for a career in the local steelworks. I chose computer science, where I saw a demonstration of an early artificial intelligence (AI) program that analysed written English. From then on, I was hooked on the idea of AI.
I worked in computer vision for many years, developing techniques to extract information from images. One day, someone came into my office and told me about a biologist who wanted to bring mathematical modelling to plant science. The problem was that biologists produced microscope images and mathematicians needed numbers – I was the glue between them. While working on that project, it became apparent that phenotyping was holding back advances in crop improvement.
After 15 years developing plant phenotyping methods, I have a good understanding of the problems faced by plant scientists, but I’m not a plant scientist. I am one of the few phenotyping researchers who came to the field from the computer science side (most are biologists who become interested in computing), giving me a better understanding of what is technologically possible. PhenomUK is about bringing computer scientists and engineers into plant phenotyping, and I was well-placed to do that.
I don’t actually know very much about plants – I’m still a computer scientist at heart. For example, I was surprised to learn that when pests attack a crop, some plants react by releasing chemicals through their roots. Nearby plants pick these up through their roots and produce a kind of insect repellent through their leaves, thereby protecting the rest of the field.
MEET CLAIRE

CLAIRE HAYES IS THE NETWORK MANAGER OF PHENOMUK
I loved horses when I was younger and wanted to be a veterinary nurse. I worked at a local vets after leaving school, but realised I needed to continue my education so I went back to college to do my A-levels.
I then studied biomedical science at university and worked in pathology when I graduated. After having children, I wanted a part-time position so came back to work at the university in administration, first in the Faculty of Engineering and now as the Network Manager for PhenomUK.
My work involves everything from branding and webpage design to arranging meetings and events. I manage our social media accounts, collaborate with other academics and institutions, host online workshops and oversee the application process for research funding.
Good communication skills are key for my role. It can be challenging to keep on top of all the tasks, so being pro-active and organised is also essential.
I love the variability of my job. Some days I’ll be promoting the network on social media, other days I’ll be planning workshops. Phenotyping has such a wide scope of interesting topics so it’s a great subject to be involved with, and I enjoy hearing about the interesting research happening in the field.
I love the fact that plants and humans are uniquely connected by the photosynthesis and respiration process. As we breath out carbon dioxide, plants take it in and provide us with oxygen. That is why we must take care of nature – so it can take care of us.
TONY’S TOP TIP
Develop your computer coding skills and take an interest in real-world challenges that plant phenotyping can help address.
Do you have a question for Tony?
Write it in the comments box below and Tony will get back to you. (Remember, researchers are very busy people, so you may have to wait a few days.)