Saving plants from disease
To prevent outbreaks of disease destroying wheat and other vital food crops, scientists from the Rothamsted Research and European Bioinformatics Institutes are sharing genetic and genomic information on disease-causing microorganisms
Every year, up to one-quarter of plants grown for food are spoiled or killed outright by disease before the crop can be harvested. Those that survive are sometimes later destroyed by disease while being transported around the world. It’s hard to predict where in the world new outbreaks will happen. As the world warms up due to climate change, the microorganisms (fungi, protists, bacteria and viruses) that cause diseases are gradually moving towards Earth’s poles. They can also be carried to new locations by severe storms, and inadvertently by humans, animals, or traded goods. So, what can be done to solve this problem?
To prevent plant diseases, scientists at the Rothamsted Research and European Bioinformatics Institutes are sharing detailed genetic and genomic information about ‘pathogenic organisms’ contained in two databases: PHI-base and PhytoPath.
WHAT ARE THE PATHOGEN-HOST INTERACTIONS DATABASE (PHI-BASE) AND PHYTOPATH?
PHI-base and PhytoPath are databases that help researchers discover more about organisms that cause disease (plant pathogens). Scientists use these databases to look up information on a pathogen to find out which genes and gene clusters are responsible for infecting a plant (the host). This improved knowledge is important for devising ways to prevent diseases from occurring in the first place, or to reduce the spread of disease because a disease may go on to infect either more plants of the same species, or other plant species.
Genes provide the instructions that tell plants, animals and humans how to make proteins, including enzymes and chemical messengers. In humans, for example, a variety of genes determine the colour of your eyes or hair, and we inherit different genes from each of our parents depending on the genetic segregation battles that take place in specific cells. All of the genes in a given organism make up its genome (its DNA blueprint), and it’s the DNA sequence of genes that govern the proteins, enzymes and chemical messengers that are produced. The genes known to cause pathogenic diseases are included in PHI-base. These can be accessed from PhytoPath, as well as whole genome sequences that it pulls in from another database called Ensembl.
PHI-base is updated regularly, with the latest version including data on 6,438 genes from 263 pathogens and 194 hosts. It provides detailed information on the genes that affect the outcome of interactions between pathogens and hosts. It also gives information on ‘host target sites’, which detect the presence of a pathogen and activate the plant’s defences.
WHAT KIND OF ORGANISMS CAUSE PLANT DISEASES?
PHI-base and PhytoPath include data on three types of plant pathogen: fungi, bacteria, and single-celled organisms called protists. For example, a fungus that causes stem rust is called Puccinia graminis; a bacterium that stops rice seeds from growing is called Burkholderia glumae; and a protist that causes potato blight is called Phytophthora infestans.
Too small to be seen with the naked eye, protists are sometimes called water moulds. That’s because many species can swim in water, thanks to a microscopic whip-like appendage called a flagellum. Many protists are dispersed and infect plants only when it rains. As well as infecting plants, protists are also responsible for major diseases in humans and animals, including malaria.
Strange as it may seem, not all diseases caused by pathogens are undesirable. The Botrytis cinerea fungus causes grey mould disease on a large number of fruit and vegetable species, which is definitely unwanted. But it is also responsible for ‘noble rot’, which wine producers love. Grapes that pick up this infection at the right time produce fine, concentrated sweet wine.
WHO USES THESE DATABASES?
Biologists, molecular geneticists and molecular biologists use PHI-base. They can find out how a variation in a gene, called a mutation, affects its characteristics. For example, a mutation might make the pathogen more or less virulent (harmful). Or scientists may have an interest in a specific phenotype (appearance or trait) that they wish to find out more about.
Other scientists, known as bioinformaticians and computational biologists, use PHI-base and PhytoPath to study organisms using computers. They compare different species, identifying the genes or sets of genes linked to a particular pathogen or host. Before PHI-base and PhytoPath, it would have taken months for scientists to get hold of this combined genetic and phenotypic data.
HOW WILL PHI-BASE AND PHYTOPATH HAVE THE BIGGEST IMPACT?
Plant pathogens are a major problem for agriculture around the world. In the case of a new disease outbreak, these two databases will provide valuable information so that governments and industries can act quickly to stop diseases spreading. Once a disease-causing organism has been analysed, information on its genes and genome will be available within a few days to weeks of any outbreak.
One way of stopping diseases killing or disabling plants is to spray them with pesticides. Thanks to detailed genetic and genomic data, manufacturers of anti-infectives will be able to make them more effective. Prevention is an even better way of stopping diseases. Plant breeders can exploit the data in PHI-base to create new, more resilient crops. They’ll be able to learn more about how the genes and proteins of a host can detect a pathogen’s presence and activate its defences.
CAN STUDYING PLANT DISEASES IMPROVE HUMAN HEALTH?
Yes. Alternaria alternata causes a disease in plants called leaf spot and has been known to cause upper respiratory tract infections and asthma in humans. It’s just one of many microscopic fungi with the ability to infect plants as well as animals and humans. We know far less about these ‘hidden killers from the fungal world’ because we have studied them far less than diseases caused by bacteria or transmitted by insect vectors. Sometimes such diseases can escalate, infecting several members of the same family or even spreading to different countries. Because the diseases caused by these pathogens spread in similar ways in plants and humans, we can learn a lot by comparing different species.
WHO ELSE IS INVOLVED IN THE PROJECT?
The scientists working on PHI-base and Phytopath are based at the Rothamsted Research Institute in Hertfordshire, UK, and the European Bioinformatics Institute in Cambridge, which is part of EMBL, Europe’s flagship laboratory for the life sciences. They collaborate with scientists working at the University of Cambridge and also with data specialists at Molecular Connections in Bangalore, India. The University of Cambridge scientists are part of a global research community called PomBase working on the model fungal fission yeast species. The PomBase team is helping to develop a new tool – PHI-Canto – to allow scientists to enter new information in PHI-base. Molecular Connections, meanwhile, reads and extracts the required information from specialist life science publications (using a method called information curation / biocuration) and also updates the interface to make PHI-base easier to search.
WHAT ARE THE CHALLENGES INVOLVED IN THIS KIND OF RESEARCH?
The amount of data that is being generated across the life sciences has grown enormously in recent years. One estimate is that the amount of genomic data will double every seven months! In 2020, data will be generated a million times faster than it was less than 10 years earlier. The task of storing and transmitting all this data is too big for one country alone to handle. That’s why a European organisation has launched a ‘big data’ project called ‘ELIXIR-Data for life’ to collate all this information and make it available so that scientists across Europe can use it to make discoveries.
ELIXIR will link all the databases together so that you can use one interface to look up genetic and genomic information anywhere – much like a web browser can find information held on different computers. PHI-base supplies agrigenomics data (agriculture-related genetic information) to ELIXIR. In total, 23 countries are members of ELIXIR and, in the UK, 15 universities and research organisations are involved in the project.
WHAT DOES THE FUTURE HOLD?
Over 70% of the genes predicted in the genome of every pathogenic species have no known function. There is a lot that remains to be discovered before we fully understand how pathogens cause disease and how host species stop infections.
Five deadly plant pathogens
 PHYTOPHTHORA INFESTANS: This protist caused the world’s most deadly potato disease. Between 1845 and 1851, it was responsible for the Irish Potato Famine, which led to the deaths of 21% of the Irish population. This is still the number one potato disease, globally.
PHYTOPHTHORA INFESTANS: This protist caused the world’s most deadly potato disease. Between 1845 and 1851, it was responsible for the Irish Potato Famine, which led to the deaths of 21% of the Irish population. This is still the number one potato disease, globally.
 MAGNAPORTHE ORYZAE: This fungus causes rice blast disease – the world’s number one disease affecting rice. In some parts of the world, its close cousin now infects wheat.
MAGNAPORTHE ORYZAE: This fungus causes rice blast disease – the world’s number one disease affecting rice. In some parts of the world, its close cousin now infects wheat.
 PUCCINIA FUNGAL RUST FAMILY: The individual species in this family specialise in causing diseases in specific cereals. For example, black stem rust disease on wheat is a major problem for agriculture, which in the 1950s completely destroyed the USA wheat crop for several years.
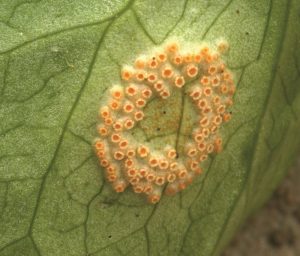
PUCCINIA FUNGAL RUST FAMILY: The individual species in this family specialise in causing diseases in specific cereals. For example, black stem rust disease on wheat is a major problem for agriculture, which in the 1950s completely destroyed the USA wheat crop for several years.
 FUSARIA FUNGAL FAMILY: The species in this family affect cereal crops as well as bananas, tomatoes and vegetables. They can also cause illness in humans, including eye disease and, in some people, diseases of the lungs and blood. These fungi are also able to live in the soil and on decaying plant tissue once a crop is harvested.
FUSARIA FUNGAL FAMILY: The species in this family affect cereal crops as well as bananas, tomatoes and vegetables. They can also cause illness in humans, including eye disease and, in some people, diseases of the lungs and blood. These fungi are also able to live in the soil and on decaying plant tissue once a crop is harvested.
 COLLETOTRICHUM KAHAWAE: This fungus causes coffee berry disease. In 1869, it devastated so many coffee plantations in Ceylon (now Sri Lanka) that farmers switched to growing tea. Tea leaves were exported to Britain, creating a nation of tea lovers in the process!
COLLETOTRICHUM KAHAWAE: This fungus causes coffee berry disease. In 1869, it devastated so many coffee plantations in Ceylon (now Sri Lanka) that farmers switched to growing tea. Tea leaves were exported to Britain, creating a nation of tea lovers in the process!
Reference
https://doi.org/10.33424/FUTURUM12
Joint PhytoPath-PHI-base team meeting at the Hinxton Genome Campus, with the new ELIXIR building at the back left. Team members from left to right are: Uma Maheswari, Helder Pedro, Alayne Cuzick, Nishadi De Silva, Paul Kersey, Kim Hammond-Kosack and Martin Urban.
PHI-base team members based at Rothamsted Research.
Phytophthora infestans
Fusaria Fungal Family
Magnaporthe Oryzae
Colletotrichum Kahawae
Every year, up to one-quarter of plants grown for food are spoiled or killed outright by disease before the crop can be harvested. Those that survive are sometimes later destroyed by disease while being transported around the world. It’s hard to predict where in the world new outbreaks will happen. As the world warms up due to climate change, the microorganisms (fungi, protists, bacteria and viruses) that cause diseases are gradually moving towards Earth’s poles. They can also be carried to new locations by severe storms, and inadvertently by humans, animals, or traded goods. So, what can be done to solve this problem?
To prevent plant diseases, scientists at the Rothamsted Research and European Bioinformatics Institutes are sharing detailed genetic and genomic information about ‘pathogenic organisms’ contained in two databases: PHI-base and PhytoPath.
WHAT ARE THE PATHOGEN-HOST INTERACTIONS DATABASE (PHI-BASE) AND PHYTOPATH?
PHI-base and PhytoPath are databases that help researchers discover more about organisms that cause disease (plant pathogens). Scientists use these databases to look up information on a pathogen to find out which genes and gene clusters are responsible for infecting a plant (the host). This improved knowledge is important for devising ways to prevent diseases from occurring in the first place, or to reduce the spread of disease because a disease may go on to infect either more plants of the same species, or other plant species.
Genes provide the instructions that tell plants, animals and humans how to make proteins, including enzymes and chemical messengers. In humans, for example, a variety of genes determine the colour of your eyes or hair, and we inherit different genes from each of our parents depending on the genetic segregation battles that take place in specific cells. All of the genes in a given organism make up its genome (its DNA blueprint), and it’s the DNA sequence of genes that govern the proteins, enzymes and chemical messengers that are produced. The genes known to cause pathogenic diseases are included in PHI-base. These can be accessed from PhytoPath, as well as whole genome sequences that it pulls in from another database called Ensembl.
PHI-base is updated regularly, with the latest version including data on 6,438 genes from 263 pathogens and 194 hosts. It provides detailed information on the genes that affect the outcome of interactions between pathogens and hosts. It also gives information on ‘host target sites’, which detect the presence of a pathogen and activate the plant’s defences.
WHAT KIND OF ORGANISMS CAUSE PLANT DISEASES?
PHI-base and PhytoPath include data on three types of plant pathogen: fungi, bacteria, and single-celled organisms called protists. For example, a fungus that causes stem rust is called Puccinia graminis; a bacterium that stops rice seeds from growing is called Burkholderia glumae; and a protist that causes potato blight is called Phytophthora infestans.
Too small to be seen with the naked eye, protists are sometimes called water moulds. That’s because many species can swim in water, thanks to a microscopic whip-like appendage called a flagellum. Many protists are dispersed and infect plants only when it rains. As well as infecting plants, protists are also responsible for major diseases in humans and animals, including malaria.
Strange as it may seem, not all diseases caused by pathogens are undesirable. The Botrytis cinerea fungus causes grey mould disease on a large number of fruit and vegetable species, which is definitely unwanted. But it is also responsible for ‘noble rot’, which wine producers love. Grapes that pick up this infection at the right time produce fine, concentrated sweet wine.
WHO USES THESE DATABASES?
Biologists, molecular geneticists and molecular biologists use PHI-base. They can find out how a variation in a gene, called a mutation, affects its characteristics. For example, a mutation might make the pathogen more or less virulent (harmful). Or scientists may have an interest in a specific phenotype (appearance or trait) that they wish to find out more about.
Other scientists, known as bioinformaticians and computational biologists, use PHI-base and PhytoPath to study organisms using computers. They compare different species, identifying the genes or sets of genes linked to a particular pathogen or host. Before PHI-base and PhytoPath, it would have taken months for scientists to get hold of this combined genetic and phenotypic data.
HOW WILL PHI-BASE AND PHYTOPATH HAVE THE BIGGEST IMPACT?
Plant pathogens are a major problem for agriculture around the world. In the case of a new disease outbreak, these two databases will provide valuable information so that governments and industries can act quickly to stop diseases spreading. Once a disease-causing organism has been analysed, information on its genes and genome will be available within a few days to weeks of any outbreak.
One way of stopping diseases killing or disabling plants is to spray them with pesticides. Thanks to detailed genetic and genomic data, manufacturers of anti-infectives will be able to make them more effective. Prevention is an even better way of stopping diseases. Plant breeders can exploit the data in PHI-base to create new, more resilient crops. They’ll be able to learn more about how the genes and proteins of a host can detect a pathogen’s presence and activate its defences.
CAN STUDYING PLANT DISEASES IMPROVE HUMAN HEALTH?
Yes. Alternaria alternata causes a disease in plants called leaf spot and has been known to cause upper respiratory tract infections and asthma in humans. It’s just one of many microscopic fungi with the ability to infect plants as well as animals and humans. We know far less about these ‘hidden killers from the fungal world’ because we have studied them far less than diseases caused by bacteria or transmitted by insect vectors. Sometimes such diseases can escalate, infecting several members of the same family or even spreading to different countries. Because the diseases caused by these pathogens spread in similar ways in plants and humans, we can learn a lot by comparing different species.
WHO ELSE IS INVOLVED IN THE PROJECT?
The scientists working on PHI-base and Phytopath are based at the Rothamsted Research Institute in Hertfordshire, UK, and the European Bioinformatics Institute in Cambridge, which is part of EMBL, Europe’s flagship laboratory for the life sciences. They collaborate with scientists working at the University of Cambridge and also with data specialists at Molecular Connections in Bangalore, India. The University of Cambridge scientists are part of a global research community called PomBase working on the model fungal fission yeast species. The PomBase team is helping to develop a new tool – PHI-Canto – to allow scientists to enter new information in PHI-base. Molecular Connections, meanwhile, reads and extracts the required information from specialist life science publications (using a method called information curation / biocuration) and also updates the interface to make PHI-base easier to search.
WHAT ARE THE CHALLENGES INVOLVED IN THIS KIND OF RESEARCH?
The amount of data that is being generated across the life sciences has grown enormously in recent years. One estimate is that the amount of genomic data will double every seven months! In 2020, data will be generated a million times faster than it was less than 10 years earlier. The task of storing and transmitting all this data is too big for one country alone to handle. That’s why a European organisation has launched a ‘big data’ project called ‘ELIXIR-Data for life’ to collate all this information and make it available so that scientists across Europe can use it to make discoveries.
ELIXIR will link all the databases together so that you can use one interface to look up genetic and genomic information anywhere – much like a web browser can find information held on different computers. PHI-base supplies agrigenomics data (agriculture-related genetic information) to ELIXIR. In total, 23 countries are members of ELIXIR and, in the UK, 15 universities and research organisations are involved in the project.
WHAT DOES THE FUTURE HOLD?
Over 70% of the genes predicted in the genome of every pathogenic species have no known function. There is a lot that remains to be discovered before we fully understand how pathogens cause disease and how host species stop infections.
Five deadly plant pathogens
 PHYTOPHTHORA INFESTANS: This protist caused the world’s most deadly potato disease. Between 1845 and 1851, it was responsible for the Irish Potato Famine, which led to the deaths of 21% of the Irish population. This is still the number one potato disease, globally.
PHYTOPHTHORA INFESTANS: This protist caused the world’s most deadly potato disease. Between 1845 and 1851, it was responsible for the Irish Potato Famine, which led to the deaths of 21% of the Irish population. This is still the number one potato disease, globally.
 MAGNAPORTHE ORYZAE: This fungus causes rice blast disease – the world’s number one disease affecting rice. In some parts of the world, its close cousin now infects wheat.
MAGNAPORTHE ORYZAE: This fungus causes rice blast disease – the world’s number one disease affecting rice. In some parts of the world, its close cousin now infects wheat.
 PUCCINIA FUNGAL RUST FAMILY: The individual species in this family specialise in causing diseases in specific cereals. For example, black stem rust disease on wheat is a major problem for agriculture, which in the 1950s completely destroyed the USA wheat crop for several years.
PUCCINIA FUNGAL RUST FAMILY: The individual species in this family specialise in causing diseases in specific cereals. For example, black stem rust disease on wheat is a major problem for agriculture, which in the 1950s completely destroyed the USA wheat crop for several years.
 FUSARIA FUNGAL FAMILY: The species in this family affect cereal crops as well as bananas, tomatoes and vegetables. They can also cause illness in humans, including eye disease and, in some people, diseases of the lungs and blood. These fungi are also able to live in the soil and on decaying plant tissue once a crop is harvested.
FUSARIA FUNGAL FAMILY: The species in this family affect cereal crops as well as bananas, tomatoes and vegetables. They can also cause illness in humans, including eye disease and, in some people, diseases of the lungs and blood. These fungi are also able to live in the soil and on decaying plant tissue once a crop is harvested.
 COLLETOTRICHUM KAHAWAE: This fungus causes coffee berry disease. In 1869, it devastated so many coffee plantations in Ceylon (now Sri Lanka) that farmers switched to growing tea. Tea leaves were exported to Britain, creating a nation of tea lovers in the process!
COLLETOTRICHUM KAHAWAE: This fungus causes coffee berry disease. In 1869, it devastated so many coffee plantations in Ceylon (now Sri Lanka) that farmers switched to growing tea. Tea leaves were exported to Britain, creating a nation of tea lovers in the process!
ASK DR KIM HAMMOND-KOSACK AND DR PAUL KERSEY
HOW DID PHI-BASE AND PHYTOPATH BEGIN?
Kim: I wanted to discover how pathogens attack plants to cause disease. Between 1999 and 2002, with the help of Dr Martin Urban, I curated all the relevant published papers into an Excel table. We realised there wasn’t a database available for the type of gene function we were interested in, so in 2003, the Excel sheet turned into the database called PHI-base.
Paul: PhytoPath came about because I was looking to connect Ensembl (a database I was already operating) to more functional data from pathogen species – and PHI-base was an obvious partner.
WHAT WAS THE KEY DISCOVERY IN YOUR FIELD OF RESEARCH?
In the early 1980s in California (USA), Doug Dahlbeck, Noel Keen and Brian Staskawicz carried out a game-changing experiment concerning bacterial avirulence genes, which trigger disease resistance during infection of a specific host plant. They used a method called ‘shotgun cloning’ of the entire genome from one avirulent race of bacteria and transferred this in small fragments into a virulent race. When the correct DNA fragment was chosen this changed the virulent race to avirulent – one that didn’t cause disease. By 1999, around 100 bacterial avirulence genes had been cloned and this data was curated into PHI-base.
WHAT’S NEXT FOR PHI-BASE AND PHYTOPATH?
We’ve developed a tool, PHI-Canto, to enable scientists to enter new information directly into PHI-base. Now we need to make videos and publicise it! We also want to catalogue the pathogen target sites of all known anti-infective chemistries for fungicides, bactericides and antibiotics, alongside the variant mutant genes that lead to unwanted microbial resistance to these chemistries.
WHO’S WHO IN THE RESEARCH TEAM
 KIM HAMMOND-KOSACK
KIM HAMMOND-KOSACK
Co-founder and team leader of PHI-base
Key skills: plant pathology, molecular plant pathology, genetics and molecular genetics, bioinformatic analysis of pathogen genomes.
Nationality: British
Why I love my job: I want to improve the health of crops, humans, animals and natural ecosystems.
Fun fact: I love photographing the natural world!
Kim has more than 30 years’ experience in molecular plant pathology and genetics, investigating the harmful fungi and viruses that infect various food plants such as wheat, tomatoes and oilseed rape. Kim and her research team work at the cutting edge with these infectious organisms – combing through their newly decoded genomes (DNA blueprints), looking for any weakness in their defences. They then share this information online with scientists who are fighting to protect their own food supplies all over the world.
 PAUL KERSEY
PAUL KERSEY
Investigator
Key skills: Bioinformatics
Nationality: British
Why I love my job: I’m interested in understanding the science, but it helps that the cause is so obviously important to mankind.
Fun fact: I was once deported from Russia!
 MARTIN URBAN
MARTIN URBAN
Co-founder of PHI-base
Key skills: Molecular plant pathology of fungal pathogens, computer sciences
Nationality: German
Why I love my job: I’m fascinated by, and really enjoy, understanding how microbial pathogens can infect plants and animals.
Fun fact: I love jazz!
 NISHADI DE SILVA
NISHADI DE SILVA
Project leader
Key skills: Bioinformatics, computer science, science communication
Nationality: British Sri Lankan
Why I love my job: The work I’m involved in is directly linked to improving health and welfare.
Fun fact: I love running!
 ALAYNE CUZICK
ALAYNE CUZICK
PHI-base biocurator
Key skills: Biocuration (organising biological data and making it accessible)
Nationality: British
Why I love my job: I have always been fascinated by nature – it’s so essential for the health of the planet.
Fun fact: I love plants – I always have at least 10 at home!
 HELDER PEDRO
HELDER PEDRO
Bioinformatician
Key skills: Genomics, computer programming, web development
Nationality: Portuguese
Why I love my job: PhytoPath helps to research crop protection in an environmentally-friendly way.
Fun fact: I’m very sarcastic!
 JAMES SEAGER
JAMES SEAGER
Software developer
Key skills: Software development and training
Nationality: British
Why I love my job: I believe in the value of sharing knowledge and using technology to simplify complicated tasks.
Fun fact: I can rapidly shake my eyeballs!
WORKING TOGETHER, THE RESEARCH TEAM HAS SKILLS IN ALL OF THE FOLLOWING FIELDS!
- Big Data
- Molecular Biology
- Molecular Genetics
- Informatics (Handling, analysing and displaying complex and large datasets)
- Bioinformatics, especially Comparative Genomics (where genome sequences of different species are compared)
- Molecular Plant Pathology
- Plant Disease Resistance / Plant Defence
- Host and Pathogen Target Site Discovery
- Effectoromics (the study of small entities produced by pathogens that are directly transferred into the host to make it unwell).
- Ontologies (English language for literature curation and the development of controlled vocabularies)
- Database Engineering / Computer Programming
WHAT ARE BIG DATA, MOLECULAR BIOLOGY AND MOLECULAR GENETICS?
BIG DATA: Modern biological experiments generate huge quantities of data. If we can organise and describe these data properly, we can look for statistically significant patterns, which can suggest mechanisms of action. Computers can even learn to draw these conclusions for themselves – this is one type of what is often called ‘artificial intelligence’. To give you an idea of the scale of the databases Kim’s team is working with, Ensembl has around 65 terabytes of data: this is equivalent to the combined storage of 254 entry-level Apple MacBooks!
MOLECULAR BIOLOGY: At the heart of living systems is the interaction of chemical compounds known as molecules. The term ‘molecular biology’ usually refers to the study of molecules involved in heritable traits, such as DNA. But its discoveries have also produced a powerful toolset for all types of biological investigation. For example, by artificially altering the DNA of a cell, we can experimentally test what happens to an organism when a gene is absent or altered.
MOLECULAR GENETICS: Molecular genetics is a subdiscipline of molecular biology, particularly focused on functional units of DNA (known as genes) and exploring the different ways genes are inherited in families and across populations.
WHO USES DATABASES LIKE PHI-BASE AND PHYTOPATH?
- Evolutionary biologists studying how plant and pathogen genomes change and adapt over space and time
- Chemists using the newly found host and pathogen target site for the discovery of new drugs
- Plant breeders using effectoromics to select the most resistant/least susceptible plants to take forward in their breeding programmes
- Molecular biologists devising speedy pathogen diagnostic tests
- Computer modellers simulating pathogen movement locally, regionally and globally, and predicting disease epidemics and larger scale pandemics in advance
- Government scientists preparing legislation to cover particularly problematic pathogens and devise suitable quarantine measures at borders
- Governments and Non-Governmental Organisations helping farmers decide what crop species to grow/not to grow because of the anticipated changes in pathogen pressures
OPPORTUNITIES IN BIG DATA, MOLECULAR BIOLOGY AND MOLECULAR GENETICS
- The range of job opportunities in these sectors is predicted to grow dramatically in the future, and one way to make yourself highly employable is to take A levels (or equivalent qualifications) in biology, maths and computer science.
- Each year, theNuffield Foundation is offering 1,000 students – who are based in the UK and in post-16 education – paid placements to work alongside scientists, engineers, technologists and mathematicians. You have to be studying a STEM subject to qualify. Why not apply to work alongside researchers such as Kim and Paul, and find out what it’s like to be a molecular biologist?
- According to My World of Work, biologists and biochemists earn on average £43,680 per year. One big plus with a career in science is that, from your early 20s, you’re paid to travel, interact with other scientists, and present your new research findings at national and international conferences and workshops.